Table of Contents
GWAS
GWAS is one of Lifelines' additional assessments and contains the genome-wide SNP data of 15.400 adult Lifelines participants derived from the Illumina CytoSNP-12v2 array.
The name GWAS might be confusing, as the assessment is not a "genome wide association study" in itself, but rather the formation of a database of SNP data that can be used, together with Lifelines phenotype data, to perform such studies.
Note that a second set of participants is genotyped in the UGLI project.
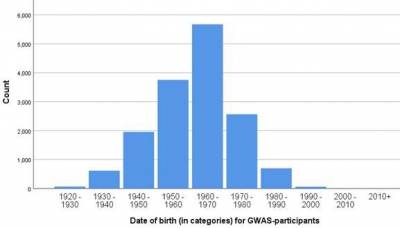
Subcohort
The GWAS subcohort consists of 15.400 independent (no biological family relations), Caucasian-ancestry adult Lifelines participants. DNA samples were collected at 1a visit 2.
Table 1: General information on the GWAS subcohort. *Age at Baseline 2nd visit.
| GWAS population general information | |
|---|---|
| Ratio male/female | 41.8% / 58.2% |
| Average age* | 47.8 |
| Minimal Age* | 18 |
| Maximum Age* | 89 |
| Age category <18 | N=0 |
| Age category 18-64 | N=13890 |
| Age category 65+ | N=1510 |
SNP array
The Illumina human CytoSNP12 Beadchip was chosen to study genetic variations in the LifeLines GWAS cohort. The 12-sample HumanCytoSNP-12 BeadChip is a powerful, whole-genome scanning panel designed for efficient, high-throughput analysis of genetic and structural variations that are the most relevant to human disease. Many types and sizes of structural variation in the human genome that affect phenotypes can be detected with the HumanCytoSNP-12 BeadChip, including duplications, deletions, amplifications, copy-neutral LOH, and mosaicism. This BeadChip includes a complete panel of genome-wide tag SNPs and markers targeting all regions of known cytogenetic importance. It incorporates 200,000 SNPs which cover around 250 genomic regions commonly screened in cytogenetics laboratories, including subtelomeric regions, pericentromeric regions, sex chromosomes, and targeted coverage in around 400 additional disease-related genes.
Quality checks
Quality controls of the data are based on SNP filtering on minor allele frequency (MAF) above 0.001, Hardy-Weinberg equilibrium (HWE) P-value >1e-4, call rate of 0.95 using Plink 1), and principal component analysis (PCA) to check for population outliers. Description and criteria for quality checks are listed in the Table 2.
Table 2: Description and criteria for quality checks
| Description | Criteria | Action |
|---|---|---|
| Hybridisation | normal range GenomeStudio from Illumina | the samples will be hybridized again |
| Call rate | call-rate < 95% | exclude data from analysis |
| Samples with call-rates < 80% are excluded before reviewing clustering | ||
| SNP genotype calling (samples with a call rate < 80% are excluded) | visual inspection of clusters for all SNPs with a GT score of < 0.51 | These clusters were changed to give the best results. After these checks a new cluster file is exported and stored with the raw data to enable exact reproduction and checking in future. This file will be used as reference cluster file in the next data release |
| Sample | ||
| Duplicate sample identification | included twice or sample mix-up? | remove data from sample with the lowest call rate |
| no relationship or mix-up can be determined | remove data from both samples | |
| Excess or deficit of heterozygote SNPs | there are consistently across the chromosomes more or less heterozygote SNPs than expected | remove data from both samples |
| there is one chromosome where there are more or less heterozygote SNPs than expected | remove data from both samples | |
| Sex check | Verify if sex is according to LifeLines database | exclude mismatches |
| Verify lab workflow to determine sex | exclude all possible mismatches | |
| SNP | ||
| Call-rate per SNP | call-rate < 95% | remove data from SNP |
| HWE equilibrium | HWE-P < 0.001 | discard for classical SNP analysis |
| Minor allele frequency | MAF <1% | exclude SNP |
Imputation
To get more information about the genome of the participants and therefore to scale up the number of SNPs, the genotype data generated using the arrays were imputed against reference genomes obtained by means of whole genome sequencing. IMPUTE2 (ref) is a program that can predict what the missing SNPs will be based on known SNPs or haplotypes (a combination of SNPs) by mapping the known genotypes against reference genomes.
Before imputation, the genotypes were pre-phased using SHAPEIT2 (ref) and aligned to the reference panels using Genotype Harmonizer (www.molgenis.org/systemsgenetics) in order to resolve strand issues. Generation of cleaned pedigree files and in- and output files for different imputation algorithms: Pedigree files are created in PLINK binary format. Genotypes of SNPs not directly genotyped on the Cyto12-SNP chip, but genotyped in another study, can be inferred by imputation. Imputation analysis is performed through Beagle 3.1.0 and data in these formats will be made available.
The samples were imputed using Minimac2) version 2012.10.3 against human reference genomes, i.e.
1000G is a human reference panel which contains world-wide collected genomes. Since only a small population of Dutch people (from the North) are part of the 1000G panel, the genotyped SNP dataset from the array was also imputed against GoNL. This is a reference panel that contains genomes from individuals with Dutch ancestry (with parents born in the Netherlands). 165 genomes in this panel were collected from Lifelines participants, meaning that genome data from Northern-Dutch people are available. The GoNL panel showed more accurate genotypes when imputing European samples compared with the 1000 Genomes Phase1 v3 reference panel. The MOLGENIS compute5) imputation pipeline was used to generate and monitor our job scripts on the distributed file system.
In summary:
- 1000G imputed: missing genotypes of participants are imputed (or predicted) based on 1000G reference panel (taken from worldwide population) and the available generated genotypes of the SNP array
- GoNL imputed: missing genotypes of participants are imputed (or predicted) based on the GoNL reference panel (taken from the Dutch population) and the available generated genotypes of the SNP array
- Unimputed: Genotypes determined on the basis of the SNP array, not imputed against genomic reference panels.
Non-Caucasian samples
To prevent false-positive association, non-Caucasian samples are excluded.
Samples are determined by:
- The LifeLines phenotype database (self-report)
- Outlier (IBS) analysis
- Population stratification (using Eigenstrat)
Cryptic relationships
Samples are selected using self reported family relations. After cleaning of the data, samples are compared with each other to determine the relationship by genetic similarity. If a pair of samples are indicated as first degree relatives, the sample with the best genotyping quality will be included.
Releasing SNP genotype data
The following files are available through the Lifelines workspace or HPC:
- files with phenotype data
- files with genotyped and imputed data
- quality control files:
- list of samples excluded
- list of SNPs excluded
- PCA component file
Abbreviations
| GWAS | Genome Wide Association Studies |
| SNP | Single-nucleotide polymorphism |
| HWE | Hardy-Weinberg Equilibrium |
| CNV | Copy Number Variant |
| MAF | Minor allele frequency |
| PLINK | PLINK is a command line program written in C/C++ |