Table of Contents
UGLI
UGLI is one of Lifelines' additional assessments. UGLI is the abbreviation for UMCG Genetics Lifelines Initiative. UGLI aims at facilitating and accelerating genetic data generation and data analysis and thereby scientific output through using the Lifelines genomics data.
Background
Genome-wide association (GWAS) data is highly valuable for biobanks such as Lifelines in identifying disease/trait associations, predicting future disease development and personalized treatment.
To facilitate the generation, analysis and study of genetic data in Lifelines, the UGLI consortium was founded. UGLI brings together many groups and PIs within the UMCG, RUG and beyond that are interested in performing such research with Lifelines data. They have brought the funding together which led to the initial genotyping of a total of 38,030 Lifelines participants, including children, as part of the HUGE consortium in Rotterdam on the Infinium Global Screening Array® (GSA) MultiEthnic Disease Version 1.0. Together with 15,400 samples already genotyped on the Cytochip (GWAS), two quality controlled GWAS datasets with a combined sample size of n~50,000 subjects will be made available to members of the UGLI consortium based on specific proposals approved by the UGLI steering committee and by Lifelines.
The UGLI consortium is actively raising funding for the genotyping of additional samples. The genotyped additional samples are generally referred to as UGLI2. With additional funding of new UGLI members, the consortium will increase the number of genotyped Lifelines participants. These efforts will make Lifelines a more interesting partner for national and international collaborations as well as with non-academic partners that work on healthy ageing.
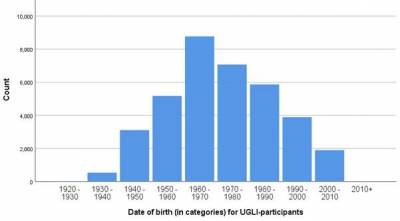
UGLI Release 1
38,030 Lifelines participants were selected for UGLI release 1 using the following criteria:
- availability of isolated DNA-samples of adequate volume and concentration at Lifelines
- Caucasian-ancestry samples
The genotype of 38,030 participants was assessed using the Infinium Global Screening Array® (GSA) MultiEthnic Disease Version 1.0. In the QC screening all genotyped samples were included, and the focuss of the QC of genetic markers was on the autosomes and chromosomes X (N=691,072 markers). A final set of 36,339 samples and 571,420 markers on autosomal and X chromosomes passed the QC steps described in QC_report_UGLI_R1.pdf.
| UGLI release 1 cohort - samples that passed QC | |
|---|---|
| Subgroup | N |
| Total | 36,339 |
| Male | 15,098 |
| Female | 21,241 |
| Age* 8-17 | 3,522* |
| Age* 18-64 | 30,416 |
| Age* >64 | 2,401 |
Table 1: UGLI release 1 cohort information. These are samples that passed QC. Age at Baseline assessment first visit. *One participant did not visit during Baseline, but did visit during 2nd screening. Since participant was under 18 years of age at 2nd screening visit 1, this participant has been added to the children 8-17 group.
Overlap between studies
| Study name | N in UGLI1 |
|---|---|
| DAG1 | ~500 |
| DAG3 | ~9000 |
| GoNL | 143* |
| GWAS4 | 938* |
Table 2: A number of participants in UGLI1 also participated in other studies, i.e. DAG1, DAG3, DAG2/GoNL and GWAS4. In the second column the sample sizes that overlap between these studies and UGLI1 can be found. For DAG1 and DAG3 these are approximations. *To evaluate sample mix-ups, a subset of samples with genotype information from a different array were compared. For this N=606 GWAS4 (CytoSNP 250k array) samples and 143 samples the Genome of the Netherlands (GoNL) project were used. See also section Quality Checks below.
SNP Genotyping Array
The Infinium Global Screening Array® (GSA) MultiEthnic Disease Version 1.0 was used for SNP genotyping of the UGLI release 1 cohort. This array contains approximately 1,000,000 SNPs and combines multi-ethnic genome-wide content, curated clinical research variants, and quality control (QC) markers for precision medicine research1).
Quality Checks
An UGLI (release 1.0) Quality Control Report is available, describing in detail the QC steps that were taken during the quality control (QC) process of the first release of UGLI comprising the genotype of 38,030 participants
assessed using the Infinium Global Screening Array® (GSA) MultiEthnic Disease Version 1.0. In this QC screening all genotyped samples were included, but the focus was on QC of genetic markers on the autosomes and chromosomes X (N=691,072 markers).
In brief, first translations and corrections specific from the GSA platform to a general
context of usage were made; namely, strand harmonization and removal of duplicate markers within the
array. Secondly, low quality samples and markers were carefully filtered with a two-steps
procedure of call rate thresholding. Further possible genotyping errors were assessed at the
marker level by detecting variants that deviated significantly from Hardy-Weinberg equilibrium
(HW) and at the sample level by evaluating heterozygosity. Then evaluated samples mix-ups
were evaluated in two levels: i) concordance of reported sex with sex derived from genotyping data from the X
and Y chromosomes, and ii) concordance of reported family information (Lifelines pedigree) and
thus of the expected genome sharing between relatives with the observed sharing from
genotyped data (genetic kinship). Moreso, to further evaluate sample mix-ups the
concordance of genotype calling among a subset of samples with genotype information from a
different array were compared (GWAS4: CytoSNP 250k array, n=606, from the Lifelines GWAS data set) and whole genome sequence (WGS, n=143, from the Genome of the Netherlands (GoNL) project).
Subsequently, Mendelian errors and further removal of SNPs that deviated from
HW in unrelated individuals was ascertained. Finally, population stratification was inspected by a principle
components analysis (PCA), incorporating samples from 1000 Genomes (1000G) and GoNL
projects. These summarized steps are shown in Figure 1 in QC_report_UGLI_R1.pdf, where each step is annotated together with the required input and whether the step generates a graphical output or a report. The code and detailed description of the process can be found in: https://github.com/molgenis/GAP.
For a more detailed description of the QC steps: QC_report_UGLI_R1.pdf
Releasing SNP Genotyping files
QCed genotype calls
A final set of 36,339 samples and 571,420 markers on autosomal and X chromosomes passing all QC steps described in QC_report_UGLI_R1.pdf will be released.
Imputation
A final set of 36,339 samples and 571,420 markers on autosomal and X
chromosomes passing all QC steps described in QC_report_UGLI_R1.pdf were used for genetic imputation. Genetic imputation was done through the Sanger imputation service
using the Haplotype Reference Consortium
( http://www.haplotype-reference-consortium.org ) panel. The dataset was formatted
following the instructions from the Sanger webpage
( https://www.sanger.ac.uk/science/tools/sanger-imputation-service ).
More details on imputation can found in QC_report_UGLI_R1.pdf
SNP array intensity files
Raw intensity data from the GSA will be made available to the researchers.
Sex mismatches
In UGLI, samples for which biological sex does not match registrated sex will be excluded from the main dataset, but not completely neglected (as opposed to GWAS). These samples will still undergo quality check analyses and will be made available for analyses to the researchers.
UGLI Release 2
As of March 2023, data of an additional 28,249 genotyped participants has been made available. Samples in this release, called UGLI2, were genotyped using the FinnGen Thermo Fisher Axiom® custom array.
29,166 participants were selected for UGLI 2 release and assessed using the pre mentioned array. All genotypes were included for QC screening, but the QC focussed on the the autosomes and chromosomes X for which there are N=617,715 and 22,405 markers available, respectively. A final set of 28,250 samples and 462,731 markers on autosomal and X chromosomes passed the QC steps described in QC_report_UGLI2_(release_1)-v1.pdf.
Please note that the array used for UGLI2 differs from the one used in UGLI1. Overlap in SNPs between these two arrays (GSA chip from Illumina=UGLI1 and FinnGen array from Affymetrix/ThermoFischer=UGLI2) is small, namely 1000-10000 SNPs.
UGLI-data release
UGLI data is available on the HPC (Linux environment) of the UMCG. The data will not be accessible through the Lifelines workspace. The applicant’s proposal will be reviewed by both Lifelines and the UGLI steering committee (UGLI SC).
UGLI consortium members receive temporary exclusive right to use the data for a period of 3 years, meaning that consortium members hold the first right to use the data should the non-UGLI member applicants proposed study overlap with a study from one of the UGLI consortium members. The 3-year period starts when UGLI-data is released on the UMCG cluster. After 3 years, UGLI consortium members must submit applications for the use of the data generated within UGLI via the regular Lifelines application procedure.
A non-UGLI consortium member requesting UGLI data has the following option:
- The applicant applies via the regular Lifelines application procedure. This means the applicant submits the proposal together with the dataset order using our online catalogue (https://data-catalogue.lifelines.nl/). UGLI data cannot be selected through the online catalogue. The applicant can request UGLI data by stating this in the written proposal. Current costs for data access will be charged. For more information on costs please visit our website: https://www.lifelines.nl/researcher/how-to-apply/costs-biobank-lifelines.
Abbreviations
| GWAS | Genome Wide Association Study |
| UGLI | UMCG Genetics Lifelines Initiative |
| UGLI SC | UGLI steering committee |
| GSA | Global Screening Array |
| SNP | Single-nucleotide polymorphism |
| HW | Hardy-Weinberg Equilibrium |
| WGS | Whole Genome Sequencing |
| MAF | Minor allele frequency |
| PCA | Principle Components Analysis |
| HPC | High Performance Computing |
| PLINK | PLINK is a command line program written in C/C++ |